Software
Automated scoring system for cervical dystonia videos
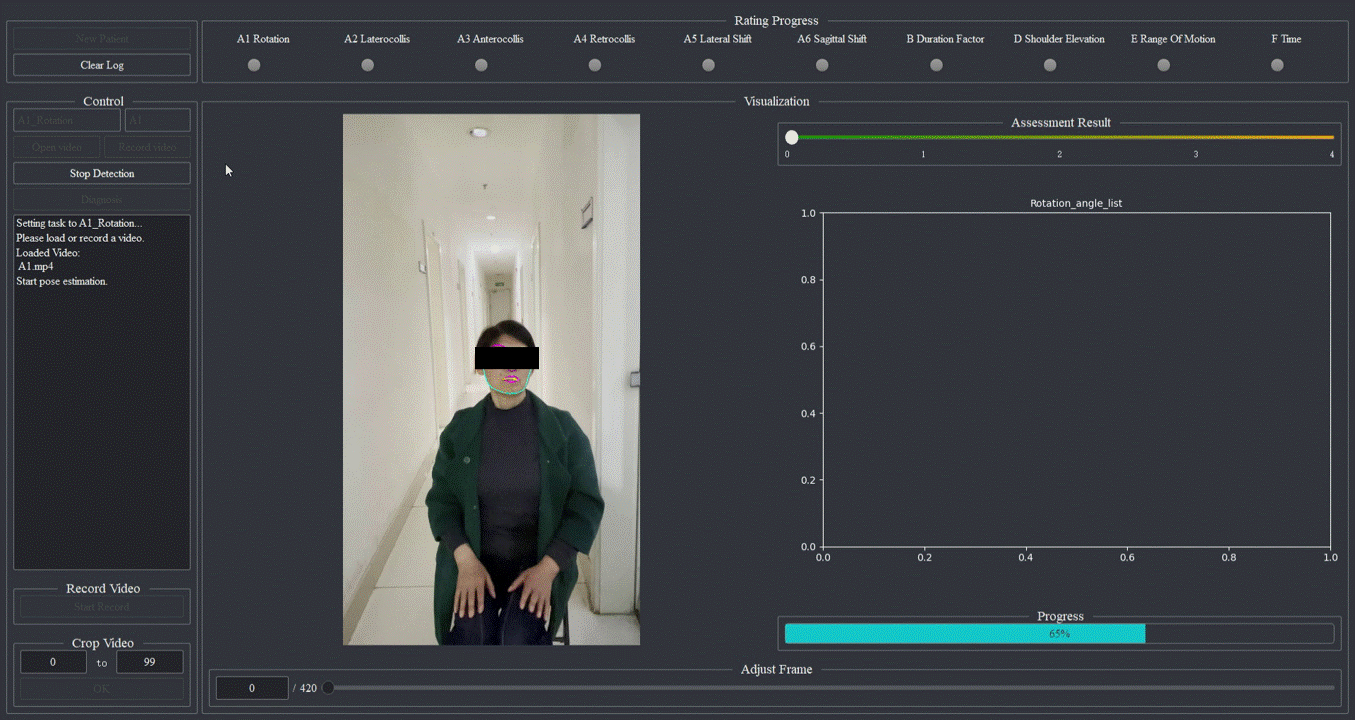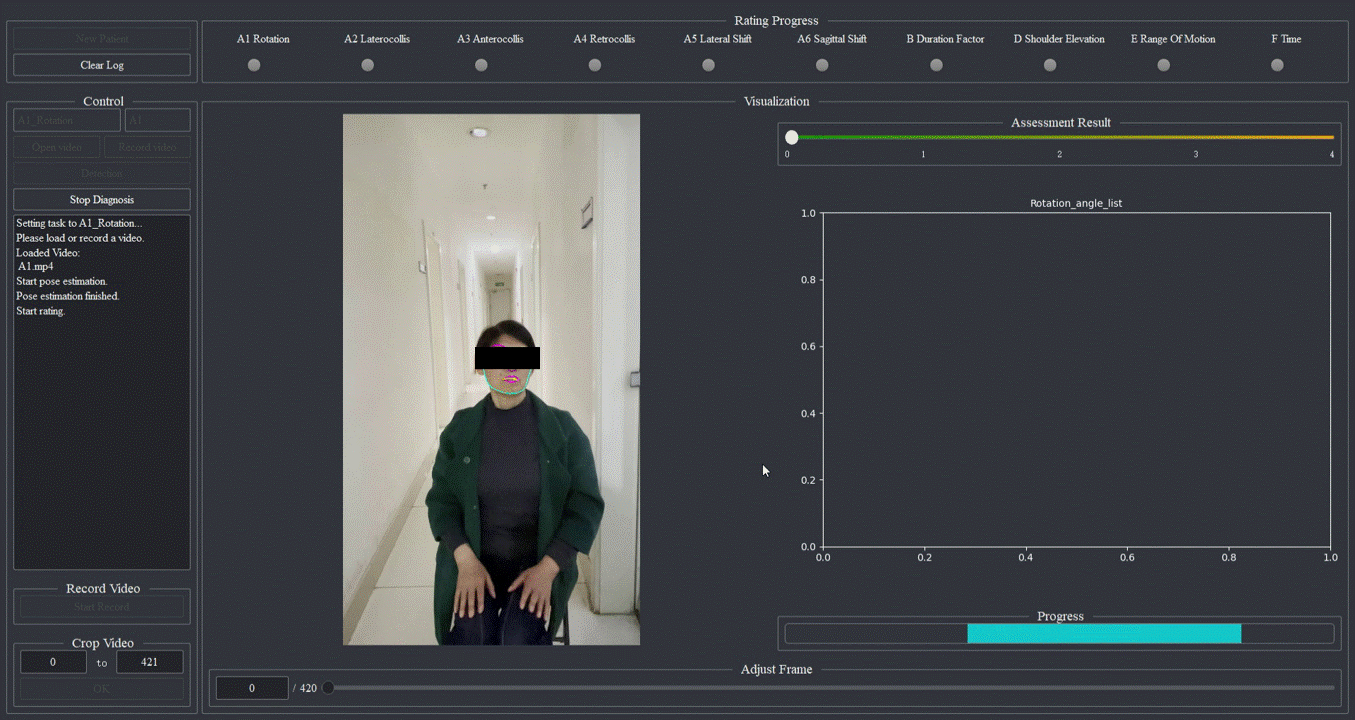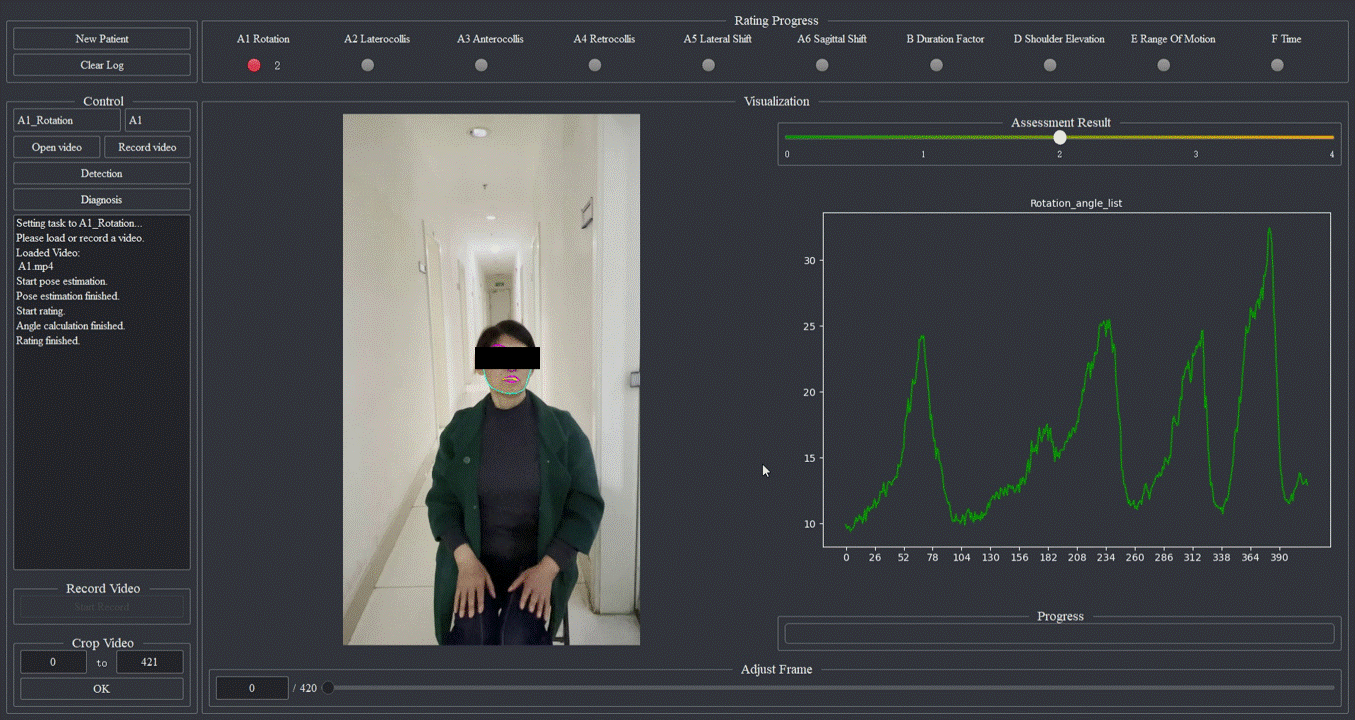
Cervical dystonia, also known as spasmodic torticollis, is a kind of movement disorder that can lead to abnormal involuntary neck muscle contraction and head posture, bringing great pain to patients. In clinical practice, the Toronto Western Spasmodic Torticollis Rating Scale (TWSTRS) is often used to assess cervical dystonia. However, there are few experienced doctors, and the visual observation assessment scheme is subjective. Therefore, we have developed an automated scoring system for cervical dystonia videos, aiming to achieve the video-based intelligent assessment of cervical dystonia through the TWSTRS. Using only the video taken by the consumer camera, the system can automatically track the patient’s face and body in the video, calculate and analyze the relevant angle parameters, and obtain the torticollis severity score in the TWSTRS, including five items: 1) maximal excursion (including six sub-items: rotation, laterocollis, anterocollis, retrocollis, lateral shift, and sagittal shift), 2) duration factor, 3) shoulder elevation, 4) range of motion, and 5) time. This low-cost system provides a valuable clinical tool for the diagnosis, assessment and even telemedicine of cervical dystonia. Currently, we are cooperating with the Department of Neurology of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine in China to carry out further clinical tests.
Automated segmentation system for pancreas and pancreatic cancer
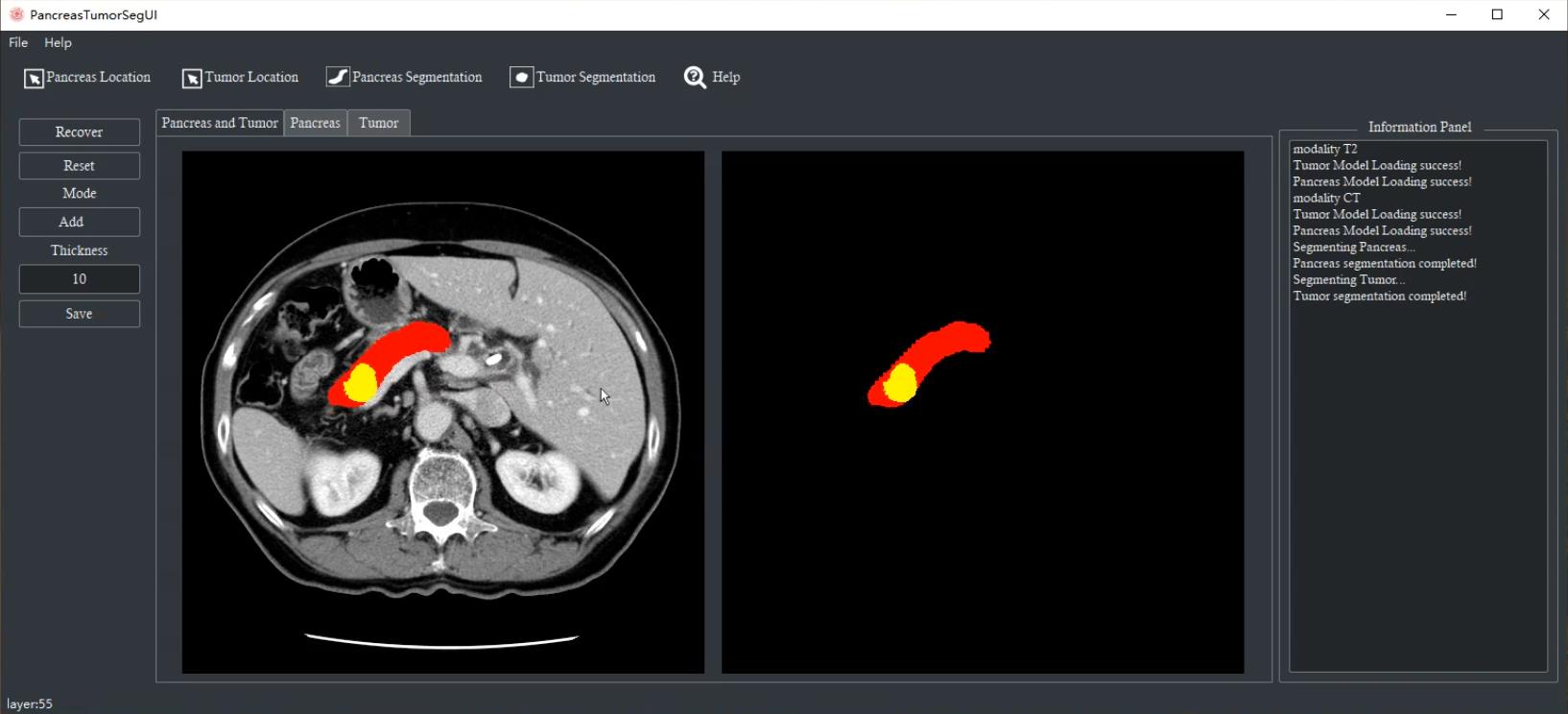
Accurate segmentation of the pancreas and pancreatic cancer is essential for various clinical studies, diagnosis and surgical treatment. However, due to the high anatomical variability, tiny volume and ambiguous borders, the pancreas and its lesions are recognized as one of the most challenging abdominal tissues for segmentation. Moreover, manual annotation is not only time-consuming and laborious, but also places high demands on the professional experience of the annotator, which will impose a huge burden on clinicians. To address this issue, we developed an automated segmentation system for pancreas and pancreatic cancer. It provides excellent generalization capabilities for fully automated segmentation on multiple modalities (such as CT and MRIs), multiple phases (such as venous-phase CT and non-contrast CT) and multi-center datasets. Moreover, we integrated the interactive functions, allowing users to update the segmentation results manually. This system is expected to provide efficient and stable tissue localization and segmentation support for pancreatic cancer clinical diagnosis and treatment (e.g., early diagnosis, surgical planning, surgical robotics, and radiotherapy).
Machine Primer Design
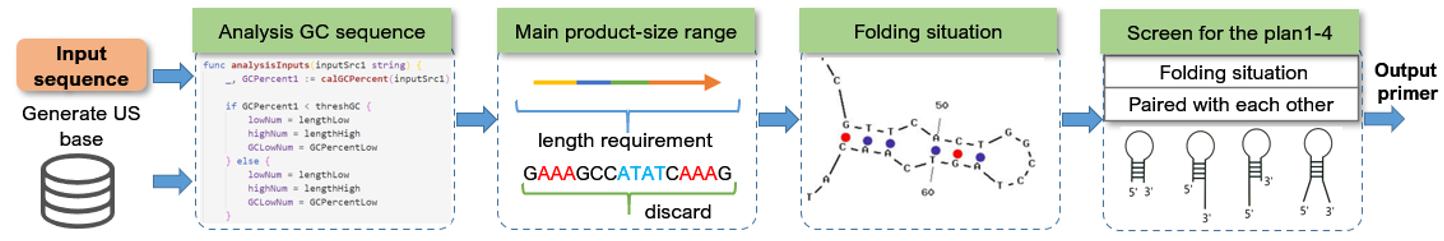
Loop-mediated isothermal amplification (LAMP) is a DNA amplification technology performed under isothermal conditions with high specificity, efficiency, and speed [Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T: Loop-mediated isothermal amplification of DNA. Nucleic Acids Research 2000, 28:E63-e63, 28(12)]. In this work, we developed an extensible program, which is designed as a flexible tool for LAMP primer design, and it can meet various design requirements in a high-throughput informatics environment. Considering the characteristics of Golang, such as high running efficiency, native high concurrency and powerful fault-tolerant mechanism, our program is completely implemented in Golang to achieve high throughput analysis based on multithreading. Besides, Golang can be deployed on each major operating system (Windows, Linux, Mac), so this program can be easily switched between different platforms.
Github RepositoryCodes
A Spatial-Transformation-Based Causality-Enhanced Model for Glioblastoma Progression Diagnosis
Qiang Li, Xinyue Li, Hong Jiang, Xiaohua Qian
PDF Github RepositoryCausality-driven candidate identification for reliable DNA methylation biomarker discovery
Xinlu Tang, Rui Guo, Zhanfeng Mo, Wenli Fu, Xiaohua Qian
PDF Github RepositoryA causal counterfactual graph neural network for arising-from-chair abnormality detection in parkinsonians
Xinlu Tang, Rui Guo, Chencheng Zhang, Xiaohua Qian
PDF Github RepositoryA Causality-Informed Graph Convolutional Network for Video Assessment of Parkinsonian Leg Agility
Rui Guo, Linbin Wang, Chencheng Zhang, Lian Gu, Dianyou Li, Xiaohua Qian
PDF Github RepositoryA Generalizable Causal-Invariance-Driven Segmentation Model for Peripancreatic Vessels
Wenli Fu, Huijun Hu, Xinyue Li, Rui Guo, Tao Chen, Xiaohua Qian
PDF Github RepositoryA Causality-Aware Graph Convolutional Network Framework for Rigidity Assessment in Parkinsonians
Xinlu Tang, Chencheng Zhang, Rui Guo, Xinling Yang, Xiaohua Qian
PDF Github RepositoryCausality-Driven Graph Neural Network for Early Diagnosis of Pancreatic Cancer in Non-Contrast Computerized Tomography
Xinyue Li, Rui Guo, Jing Lu, Tao Chen, Xiaohua Qian
PDF Github RepositoryGeneralized Pancreatic Cancer Diagnosis via Multiple Instance Learning and Anatomically-Guided Shape Normalization
Jiaqi Qu, Xunbin Wei, Xiaohua Qian
PDF Github RepositoryA Dual-transformation with Contrastive Learning Framework for Lymph Node Metastasis Prediction in Pancreatic Cancer
Xiahan Chen, Weishen Wang, Yu Jiang, Xiaohua Qian
PDF Github RepositoryGroup-shrinkage Feature Selection with a Spatial Network for Mining DNA Methylation Data
Xinlu Tang, Zhanfeng Mo, Cheng Chang, Xiaohua Qian
PDF Github RepositoryDiagnosis of Glioblastoma Multiforme Progression via Interpretable Structure-Constrained Graph Neural Networks
Xiaofan Song, Jun Li, Xiaohua Qian
PDF Github RepositoryPancreatic cancer segmentation in unregistered multi-parametric MRI with adversarial learning and multi-scale supervision
Jun Li, Chaolu Feng, Qing Shen, Xiaozhu Lin, Xiaohua Qian
PDF Github RepositoryUtilizing GCN and Meta-Learning Strategy in Unsupervised Domain Adaptation for Pancreatic Cancer Segmentation
Jun Li, Chaolu Feng, Xiaozhu Lin, Xiaohua Qian
Model-driven deep learning method for pancreatic cancer segmentation based on spiral-transformation
Xiahan Chen, Zihao Chen, Jun Li, Yu-Dong Zhang, Xiaozhu Lin, Xiaohua Qian
Auto-Metric Graph Neural Network Based on a Meta-learning Strategy for the Diagnosis of Alzheimer's disease
Xiaofan Song, Mingyi Mao, Xiaohua Qian
Combined Spiral Transformation and Model-Driven Multi-Modal Deep Learning Scheme for Automatic Prediction of TP53 Mutation in Pancreatic Cancer
Xiahan Chen, Xiaozhu Lin, Qing Shen, Xiaohua Qian
DC-AL GAN: Pseudoprogression and true tumor progression of glioblastomamultiform image classfication based on DCGAN and AlexNet
Meiyu Li,Hailiang Tang,Michael D. Chan,Xiaobo Zhou,Xiaohua Qian
Transparency-guided ensemble convolutional neural network for the stratification between pseudoprogression and true progression of glioblastoma multiform in MRI
Xiaoming Liu, Xiaobo Zhou, Xiaohua Qian
Highlights
-
One paper has been accepted by Nature Communications. Congratulations to Xinlu Tang and Rui Guo!
Jan · 2025 -
Xinlu Tang won the National Scholarships!
Sep · 2024 -
PC project team won the second prize of the National Innovation and Entrepreneurship Competition!
Aug · 2024 -
Rui Guo was honored as “Innovation Star” for Shanghai Jiao Tong University Postgraduates in 2023!
Mar · 2024 -
Pancreatic cancer screening project won the first prize at the 2023 Second Medical AI Innovation and Entrepreneurship Competition!
Dec · 2023